|
I am a PhD student at MIT advised by Vikash Mansinghka, Josh Tenenbaum, and Tim O'Donnell working on scaling data science through probabilistic programming. Previously, I was a research intern at Meta AI Research, working with Brenden Lake and Marco Baroni, at Harvard with Sam Gershman, and at the Inria Parietal team with Bertrand Thirion and Gaël Varoquaux. I've studied at École Normale Supérieure Paris-Saclay, École Polytechnique and Universidade de São Paulo. Email / CV / Google Scholar / Github |

|
|
Data science typically involves analyzing structured tables and unstructured text to make predictions, impute missing data, discover relationships between variables, infer causal effects, or detect anomalies. My work uses probabilistic programming to learn and query generative models for data science, such as guiding transformers to convert unstructured text into structured data, and learning GPU-efficient generative models for tables that can solve a wide range of data science tasks. |
|
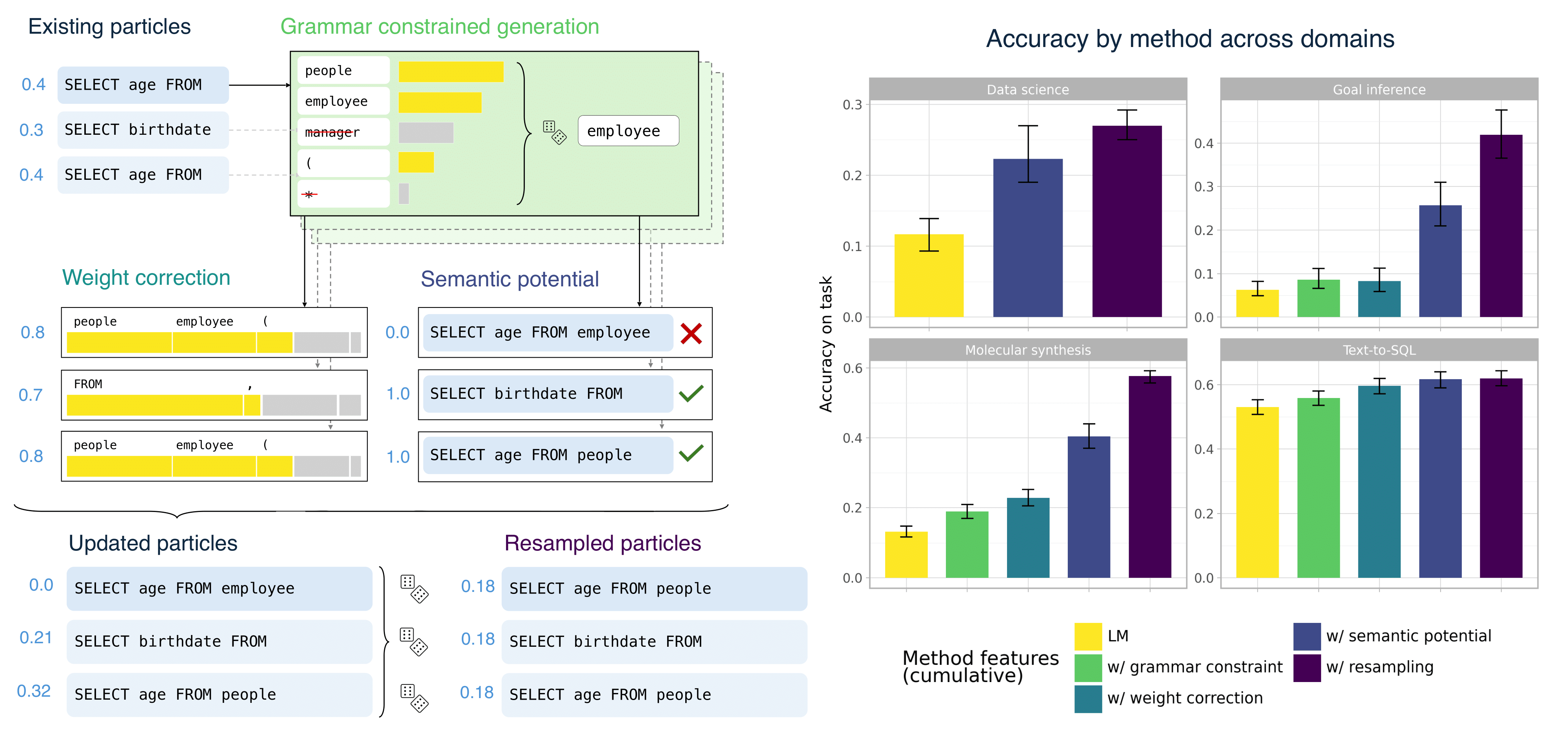
A wide range of LLM applications require generating text that conforms to syntactic or semantic constraints. Imposing such constraints nontrivially alters the distribution over sequences, usually making exact sampling intractable. In this work, building on the Language Model Probabilistic Programming framework of Lew et al. (2023), we develop an approach to approximate inference for controlled LLM generation based on sequential Monte Carlo (SMC). Our SMC framework allows us to flexibly incorporate domain- and problem-specific constraints at inference time, and efficiently reallocate computation in light of new information during the course of generation. We demonstrate that our approach improves downstream performance on four challenging domains---Python code generation for data science, text-to-SQL, goal inference, and molecule synthesis. We compare to a number of alternative and ablated approaches, showing that our accuracy improvements are driven by better approximation to the full Bayesian posterior. |
|
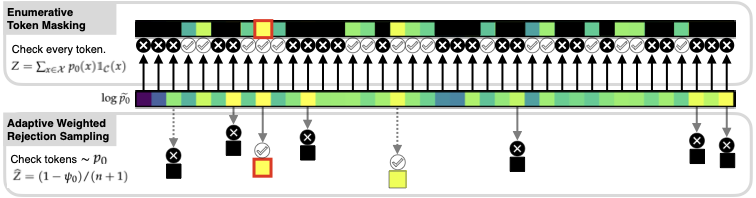
The dominant approach to generating from language models subject to some constraint is locally constrained decoding (LCD), incrementally sampling tokens at each time step such that the constraint is never violated. Typically, this is achieved through token masking: looping over the vocabulary and excluding non-conforming tokens. There are two important problems with this approach. (i) Evaluating the constraint on every token can be prohibitively expensive, since language model vocabularies often exceed 100,000 tokens. (ii) LCD can distort the global distribution over strings, sampling tokens based only on local information, even if they lead down dead-end paths. This work introduces a new algorithm that addresses both these problems. First, to avoid evaluating a constraint on the full vocabulary at each step of generation, we propose an adaptive rejection sampling algorithm that typically requires orders of magnitude fewer constraint evaluations. Second, we show how this algorithm can be extended to produce low-variance, unbiased estimates of importance weights at a very small additional cost - estimates that can be soundly used within previously proposed sequential Monte Carlo algorithms to correct for the myopic behavior of local constraint enforcement. Through extensive empirical evaluation in text-to-SQL, molecular synthesis, goal inference, pattern matching, and JSON domains, we show that our approach is superior to state-of-the-art baselines, supporting a broader class of constraints and improving both runtime and performance. Additional theoretical and empirical analyses show that our method's runtime efficiency is driven by its dynamic use of computation, scaling with the divergence between the unconstrained and constrained language model, and as a consequence, runtime improvements are greater for better models. |
|
Tabular data analysis tasks such as prediction, imputation, anomaly detection, and relationship discovery are commonplace in computational statistics, data science, and applied fields. Many popular approaches learn distinct models for every task, rather than build multivariate generative models that are guaranteed to produce coherent uncertainty measures across all queries. This paper introduces a scalable Bayesian generative modeling method for multitasking tabular data analyses, and shows that it can outperform mature, established baselines from computational statistics and deep learning. The approach learns multivariate generative models of the joint distribution over all columns in a table that provide massively parallel implementations of sampling, probability density calculation, conditioning, and marginalization. It builds on a novel GPU-accelerated, minibatch mixture Sequential Monte Carlo algorithm that exploits model structure to deliver the same asymptotic scaling as Stochastic Gradient Descent, yet produces properly weighted samples targeting the Bayesian posterior. Experiments show the approach delivers calibrated uncertainty across a broad range of dataset sizes. Experiments also show that this method can be orders of magnitude faster to train and to query than diffusion models and variational autoencoders, and outperforms task-specific methods for synthetic data generation, anomaly detection, and multiple imputation. |
|
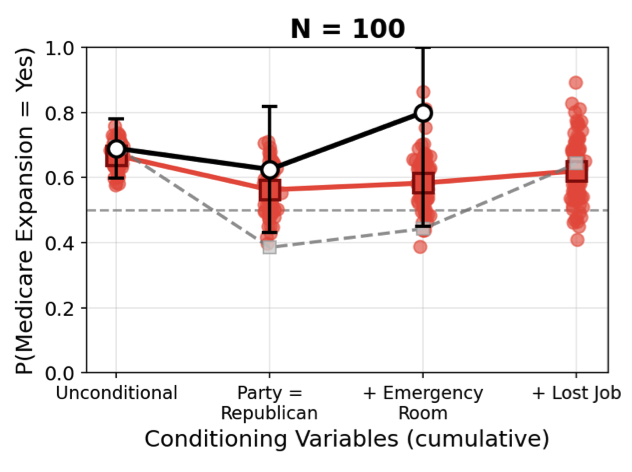
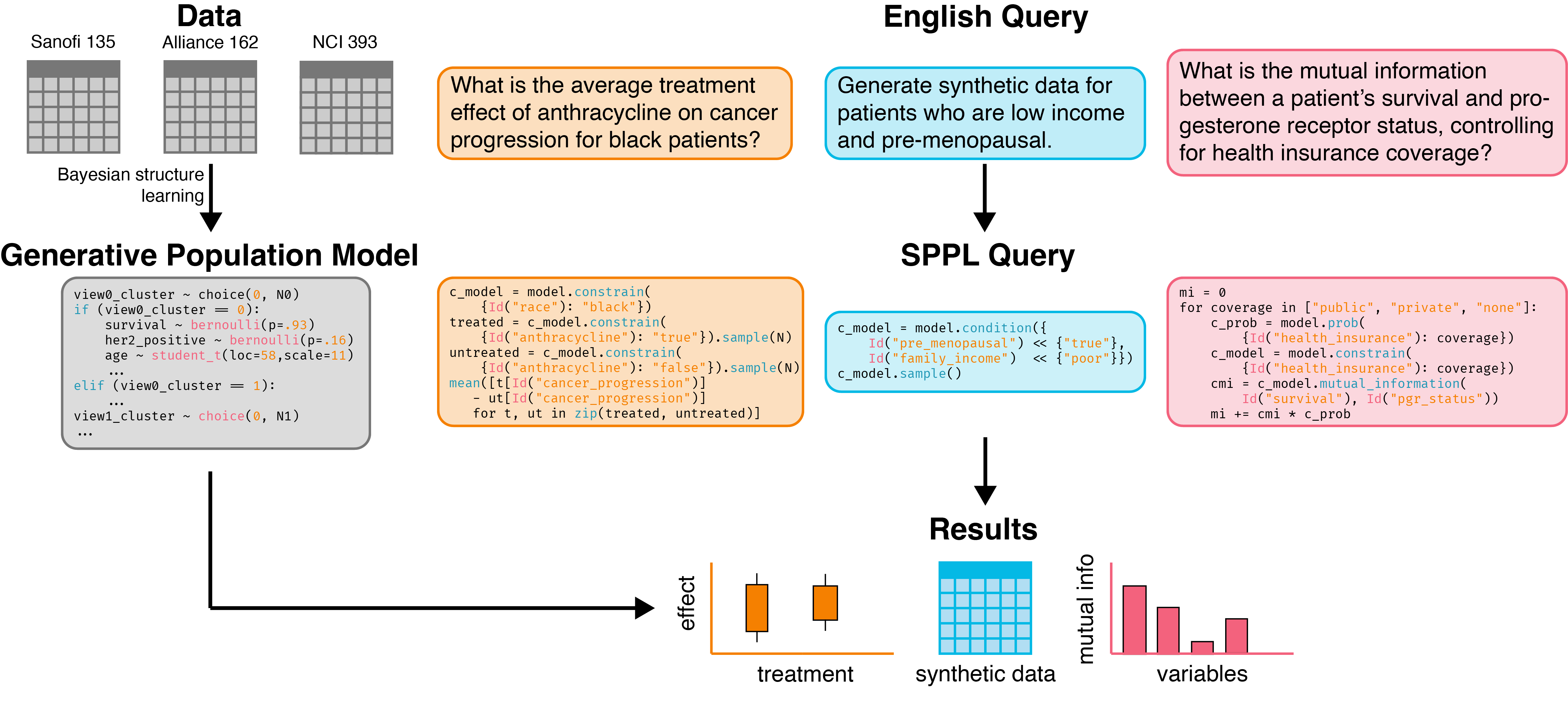
Accurate, efficient generative models of clinical populations could accelerate clinical research and improve patient outcomes. For example, such models could infer probable treatment outcomes for different subpopulations, generate high-fidelity synthetic data that can be shared across organizational boundaries, and discover new relationships among clinical variables. Using Bayesian structure learning, we show that it is possible to learn probabilistic program models of clinical populations by combining data from multiple, sparsely overlapping clinical datasets. Through experiments with multiple clinical trials and real-world evidence from census health surveys, we show that our model generates higher quality synthetic data than neural network baselines, supports more accurate inferences across datasets than traditional statistical methods, and can be queried more efficiently than both, opening up new avenues for accessible and efficient AI assistance in clinical research. |

|
Developmental psychology presents us with a puzzle: though children are remarkably apt at planning their actions, they suf- fer from surprising yet consistent shortcomings. We argue that these patterns of triumph and failure can be broadly captured by the framework of task and motion planning, where plans are hybrid entities consisting of both a structured, symbolic skeleton and a continuous, low-level trajectory. |
|
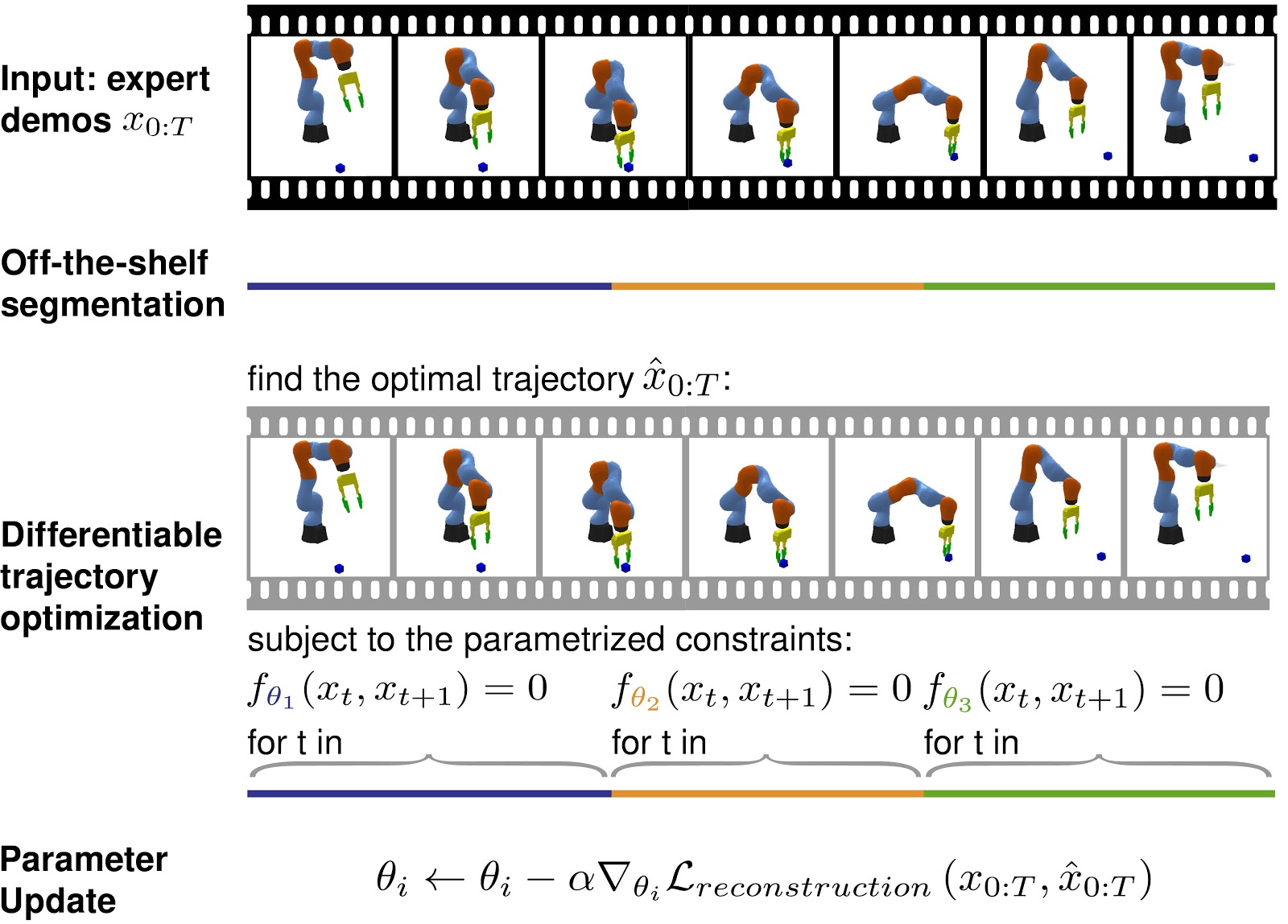
We present a framework for learning constraint-based task and motion planning models using gradient descent. Our model observes expert demonstrations of a task and decomposes them into modes—segments which specify a set of constraints on a trajectory optimization problem. |
|
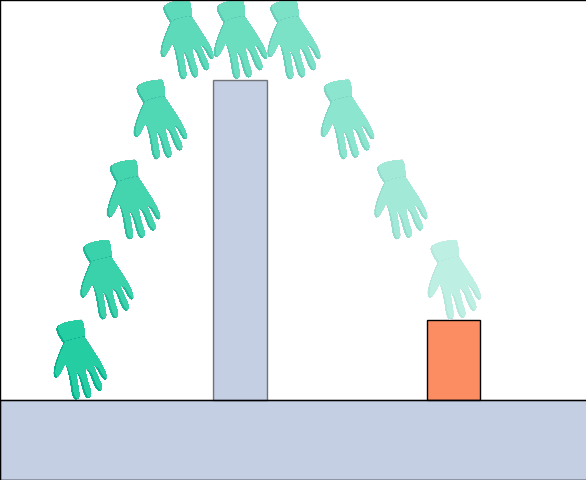
We present a model that starts out with a language of low-level physical constraints and, by observing expert demonstrations, builds up a library of high-level concepts that afford planning and action understanding. |
|
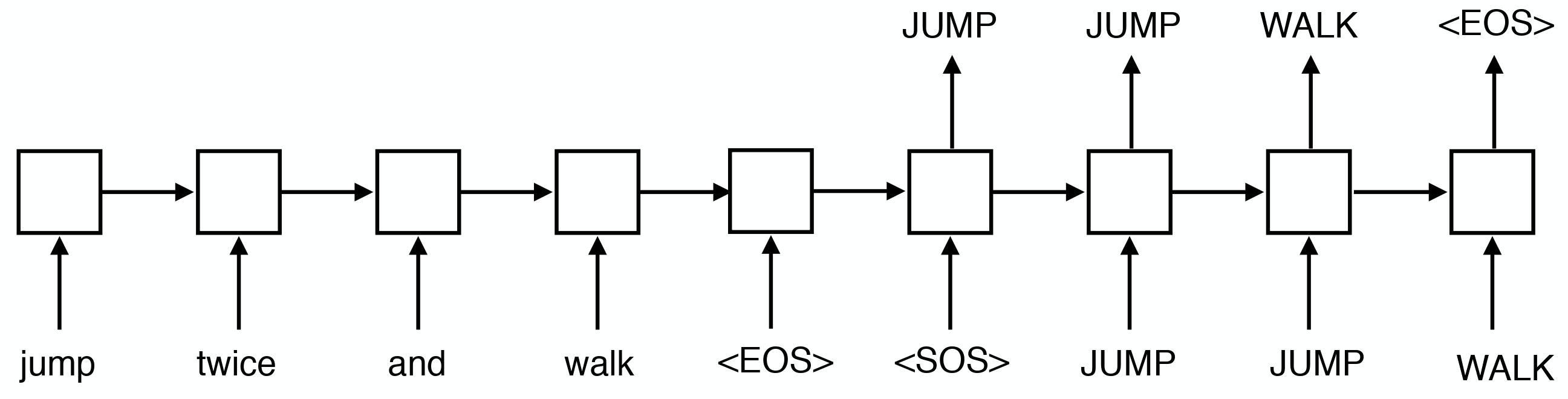
We extend the study of systematic compositionality in seq2seq models to settings where the model needs only to recombine well-trained functional words. Our findings confirm and strengthen the earlier ones: seq2seq models can be impressively good at generalizing to novel combinations of previously-seen input, but only when they receive extensive training on the specific pattern to be generalized |
|
Work on human-level learning in Atari-like games, learning theories from gameplay and using them to plan in a model-based manner. |
|
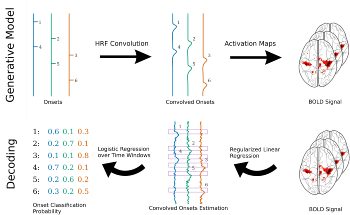
We show that fMRI decoding can be cast as a regression problem: fitting a design matrix with BOLD activation: event classification is then easily obtained from the predicted design matrices. Our experiments show this approach outperforms state of the art solutions, especially for designs with low inter-stimulus intervals, and the two-step nature of the model brings time-domain interpretability. |
|
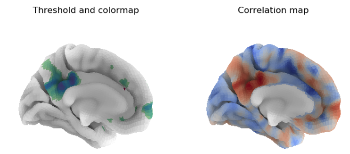
We present an initial support of cortical surfaces in Python within the neuroimaging data processing toolbox Nilearn. We provide loading and plotting functions for different surface data formats with minimal dependencies, along with examples of their application. Limitations of the current implementation and potential next steps are discussed. |
|
|